When doctors hurl toxic death at cancer cells, often a few will survive and come back. A family of enzymes called KDM5 histone demethylases is emerging as important for this resilience, and drugs that inhibit KDM5 enzymes could be active in treating several types of cancer.
A team of investigators from Emory University School of Medicine and Winship Cancer Institute, Yale, and the National Center for Advancing Translational Sciences (NCATS) of the National Institutes of Health have obtained detailed structural information, showing how inhibitors of the KDM5 family interact with their targets.
Their findings, published in Cell Chemical Biology, could inform efforts to design more potent and selective anticancer drugs.
Drugs are sometimes compared to keys, because they need to fit precisely into the enzymes they target. But there is some wiggle room, since usually more than one drug can inhibit a given enzyme. In addition, there are several families of histone demethylase enzymes, and an ongoing challenge is developing drugs that specifically inhibit KDM5 enzymes.
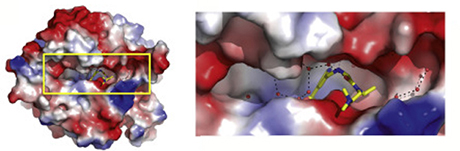
Using X-ray crystallography, scientists led by biochemists John Horton, PhD, and Xiaodong Cheng, PhD, at Emory University School of Medicine and Winship Cancer Institute, were able to see how eight different inhibitory compounds squeeze into the active site of the one member of the KDM5 family, KDM5A. In doing so, they could also see how additional potential drugs, not yet synthesized, might fit the contours of the enzyme more closely.
Scientists at NCATS identified and synthesized most of the inhibitors, several of which are being developed and tested by pharmaceutical companies. Qin Yan’s laboratory at Yale examined the growth-stopping effects of one KDM5 inhibitor on breast cancer cells.
This project, with partners at Yale and NCATS, is conducted through Emory’s Chemical Biology Discovery Center, which has been designated a Specialized Center in the Chemical Biology Consortium, the discovery engine of the National Cancer Institute’s NExT (National Cancer Institute Experimental Therapeutics) program aimed at accelerating the discovery and development of new and innovative cancer drugs through multidisciplinary research teams. Emory’s center is participating in the consortium via a research subcontract from Leidos Biomedical Research, Inc., prime contractor for the Frederick National Laboratory for Cancer Research.
What histone demethylases do and why they’re important
Histones are spool-like proteins that DNA wraps around in the cell nucleus. For a particular gene to be active and transcribed into RNA, histones have to be moved out of the way. A variety of enzymes prepare for the parade (RNA transcription) by adding signposts: chemically tweaking the histones.
KDM5 demethylases remove modifications (a –CH3 methyl group on lysine 4 of histone H3) that are a sign of a gene being active. Thus, they help maintain genes in an “off” state. KDM5 inhibitors thus are likely to keep some genes on – but they won’t necessarily act the same in all cell types.
In this paper, investigators focused on the effects on breast cancer cells, while other laboratories have shown that KDM5 enzymes are linked to drug tolerance or resistance in lung cancer, glioblastoma and melanoma.
Qin Yan’s team at Yale found that only some types of breast cancer cells’ growth is stopped by KDM5 inhibitors. For example, a cell line that was HER2+ (a marker that indicates likely response to the drug trastuzumab) is, but another cell line, representative of a “triple-negative” is not.
“We had a clue already that some breast cancer cell lines are more sensitive to KDM5 gene knockdown. This may indicate that KDM5 inhibitors will be more active toward HER2+ and luminal-type cells and less toward basal cells,” says Yan.
The research was supported by the National Cancer Institute (CA191548, CA077337 and Chemical Biology Consortium Contract HHSN261200800001E), the National Institute of General Medical Sciences (GM114306), the American Cancer Society (RSG-13-384-01-DMC), the Department of Defense (W81XWH-14-1-0308) the National Science Foundation (DGE-1122492), the Arthur and Sarah Merrill Foundation, and Winship Cancer Institute Cancer Center support grant P30-CA138292.
Xiaodong Cheng is a Georgia Research Alliance Eminent Scholar.